Physics of Cellular Systems
Research
We study the role of free energy and geometry in cell biology.
This includes how energy and geometry physically limit biological processes, and how
energy and geometry are used by cells to provide improved performance. |
| Current Projects |
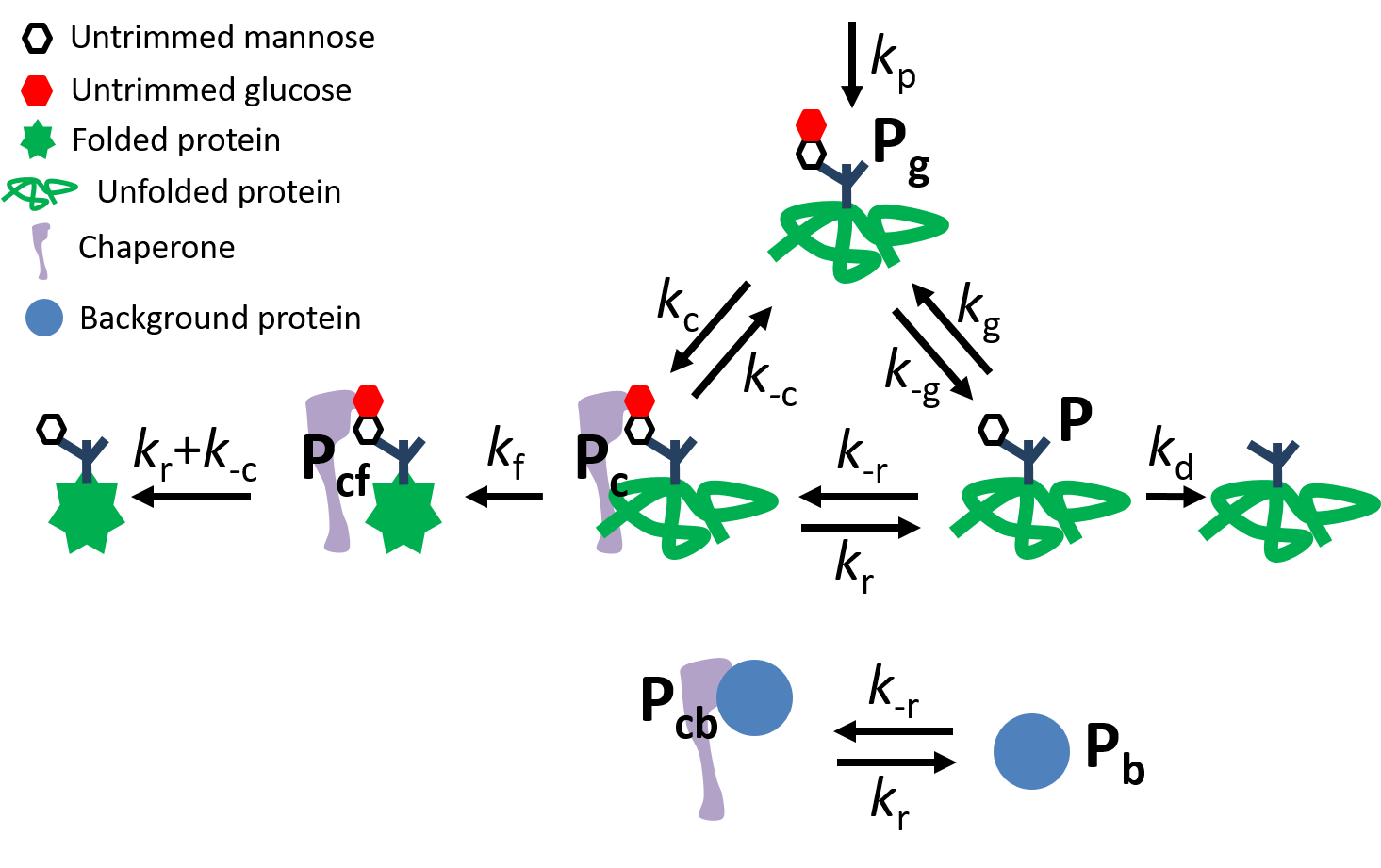 Glycoprotein quality control cycle |
Energy and kinetics for protein quality control Newly-synthesized proteins must fold into a three-dimensional structure to become functional, a
process that is often facilitated by chaperone proteins. Proteins can also misfold into an incorrect structure that is not
functional. Misfolded proteins, and proteins that have yet to fold, can aggregate, causing problems and endangering cell health.
Cells have protein 'quality control' to sort correctly folded proteins to their final functional destinations, and target misfolded
proteins for degradation. Glycoprotein quality control is a cyclic process, driven by free energy and with many kinetic steps. This
project studies how the energy use and kinetic features of glycoprotein quality control lead to improved protein production and
limited accumulation of unfolded proteins with finite chaperone available to help proteins fold. | ||
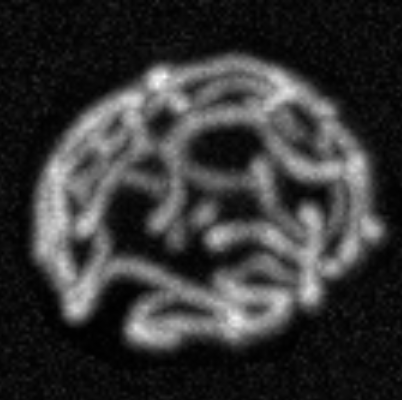 Mitochondrial network |
Mitochondrial network dynamics and protein
localization Mitochondrial tubes form extended networks with branches and loops. These networks are
dynamic, undergoing fission (tubes splitting apart), fusion (tubes merging together, both end-to-end and end-to-side), growth, and
degradation. Proteins move through these networks, diffusing, binding and unbinding, reacting, and degrading. How do network
dynamics control the spatial localization of proteins in the network? How does the impact of protein populations on network dynamics
regulate the protein populations themselves? We seek the physical limits and controlling factors for protein localization in dynamic
mitochondrial networks. | ||
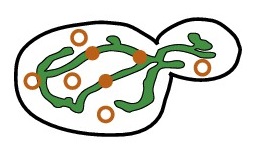 mRNA diffusing to |
Nonequilibrium control of mRNA localization to organelles
Although mitochondria have their own DNA, many mitochondrial genes are encoded in nuclear DNA, translated
into proteins by ribosomes in the cytosol, and imported into mitochondria. mRNA localization to mitochondria can play a role in
regulating mitochondrial protein concentrations. A protein can begin import into a mitochondrion as a ribosome is still translating
a protein, effectively tethering an mRNA to a mitochondrion. mRNA for some genes spend most of their time near mitochondria, some
spend very little, while others significantly increase their mitochondrial localization as the volume of mitochondria in the cell
increases. We explore how the combination of cell geometry and the nonequilibrium conditions of protein translation combine to
determine mRNA localization to mitochondria. We also probe the stochastic effects of finite mRNA number and localization on protein
concentration fluctuations between individual mitochondria. | ||
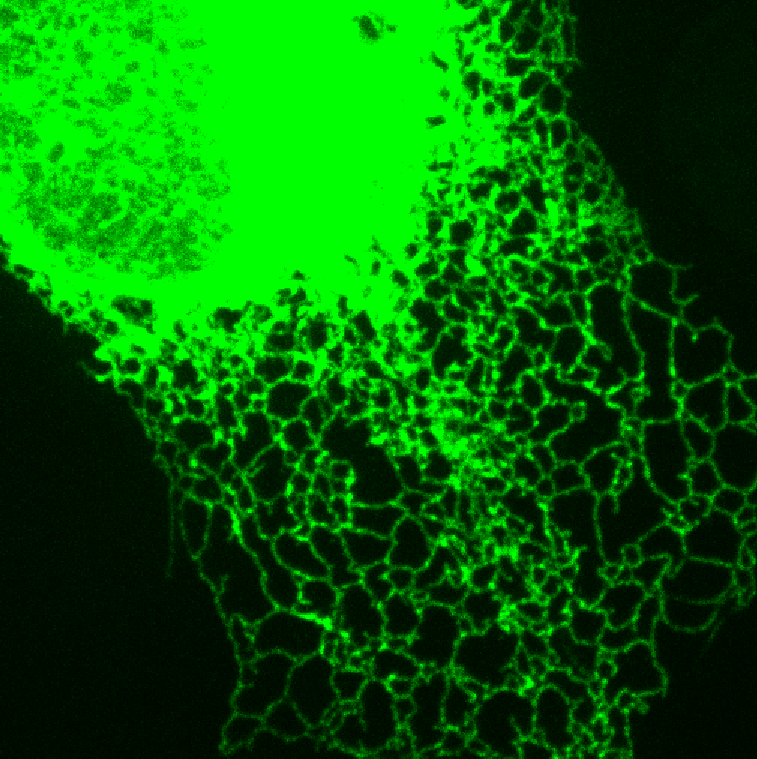 Endoplasmic reticulum network |
Biomolecular signaling dynamics under confinement Biomolecular signaling and kinetics is often understood in the relatively large volumes of a whole cell or even a test
tube. However, a lot of signaling inside cells occurs inside even smaller organellar compartments. We explore the unfolded protein
response (UPR) inside the tubes and sheets of the endoplasmic reticulum (ER). The UPR signaling system activates under the stress of
excess unfolded proteins, activating receptors in the ER membrane to form pairs and clusters. How is the behaviour of the UPR, inside
the narrow spaces within ER tubes and sheets, affected by this confinement and by the network shape of the ER? | ||
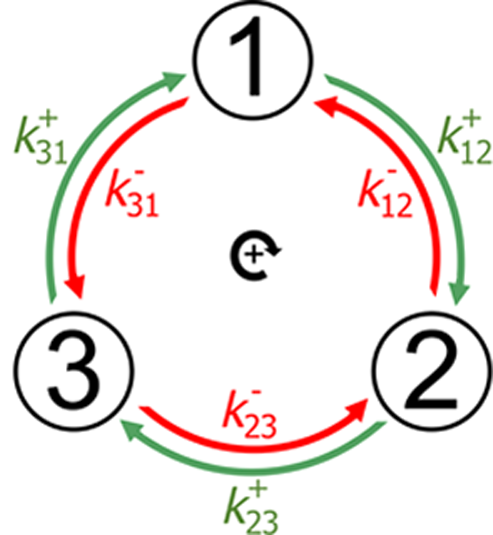 Cyclic kinetics |
Nonequilibrium selection of organelles for
degradation Cell biology is driven and directed by the dissipative expenditure of free energy. Performance
can be improved by using free energy to drive cyclic processes, such as the cycles for 'kinetic proofreading' that enable the high
accuracy of DNA replication. Another cyclic process consuming free energy is the addition of proteins that label organlles for
degradation. How does the selection of organelles for degradation improve with free energy expenditure, and trade off with other
performance measures like speed? | ||
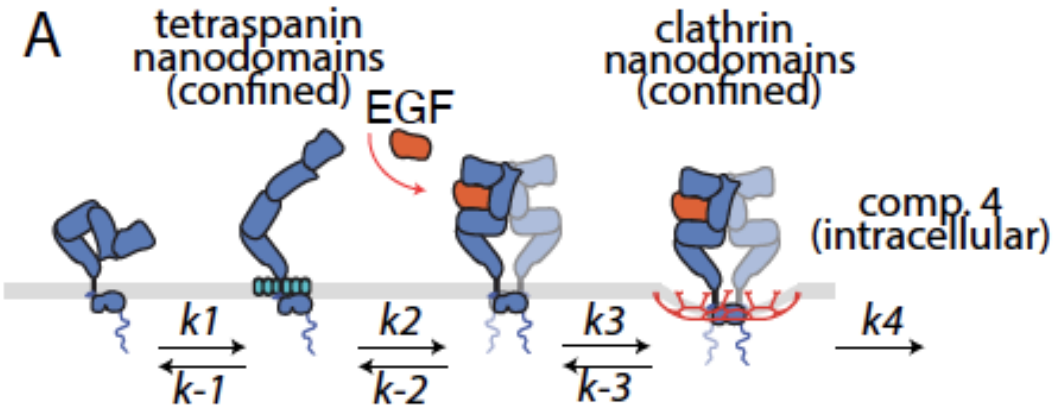 EGFR signaling state transitions |
EGFR spatial and signaling dynamics on the cell membrane
The EGF receptor (EGFR) is a cell surface receptor that is an important regulator of cell physiology and is stimulated by multiple distinct ligands.
Ligand binding to EGFR is favoured in specialized tetraspanin nanodomains on the cell surface, with ligand binding impacting EGFR dimerization, oligomerization, and post-translational modifications,
leading to downstream signaling through multiple pathways. EGFR is often internalized following ligand binding and recruitment to a clathrin domains.
Stimulation by different ligands leads to different cellular responses. How do visits to tetraspanin domains, dimerization and oligomerization dynamics, and internalization impact the time-dependent and steady-state signaling behaviour of EGFR?
|
| Published Research |